Browse this from the web or check out the generated documentation
If you have an undirected network with self loops, that is, nodes connected to themselves, the usual practice is to just drop them, eschewing thus the information they provide, whith is interesting for the evolution of the network as well as the relationship of centrality measures to status. What we propose in this package is a method that helps you take into account those edges when computing betweenness centrality.
dupNodes is the companion package to paper “Intra-family links in the analysis of marital networks”, with reference here.
It creates duplicates of nodes that have self loops in undirected graphs such as the one here
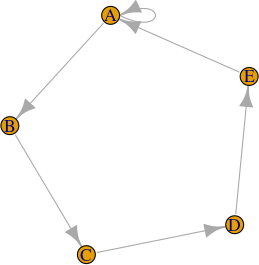
in this way
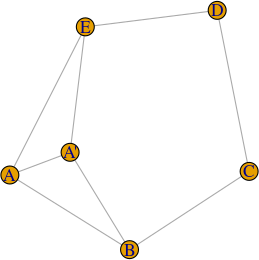
which has exactly the same degree and connections; in this way, the weights of these self-loops can be taken into account when computing centrality measures such as betweenness.
Installation
Install released versions from CRAN as usual, or development version from here
Reference
Please cite this paper when using this package in your work
@Article{mm24:intra_family_links,
author = {Merelo, J.J. and Molinari, M.C. },
title = {Intra-family links in the analysis of marital networks},
journal = {Journal of computational social science},
year = 2024,
url= {https://link.springer.com/epdf/10.1007/s42001-023-00245-4},
doi= {https://doi.org/10.1007/s42001-023-00245-4},
month = {January}
}Examples
There are some examples explained in vignettes. Use vignette("dupNodes") to access all of chem, or individually:
-
vignette("doges-social-network")to apply functions here to data from thedogesrpackage; this is the online version -
vignette("florentine-social-network"), which uses data from the well-known Florentine social network, with a (documented) self-loop added; browse it in CRAN